The famous bidomain model comes from the homogeneization of the quasi-static electric potential problem at the cell scale, where the intra- and extra-cellular potentials are defined. Assuming that the myocardium is locally constituted of a periodic pattern of cardiac cells, the bidomain problem reads:
This formulation is what is implemented in CEPS, even if it could be written in a more simple way.
# Bidomain model:
# unknown index 0 represents the transmembrane voltage
# unknown index 1 represents the extracellular potential
problem type : bidomain
#-----------------
# Output controls
#-----------------
# Output name
output file name : ./results/bidomainRL2
# Output every 0.5ms
snapshot time : 0.5
# Data used to generate the activation time map. Values are
# - Unknown index
# - AP detection threshold (mV)
# - Minimum voltage used for peak detection (mV)
# - AP duration percentage
activation time data : 0 -20 -40 0.75
# Probe points: values of the unknowns at those points*
# along time will be written in a separate file
# (*at closest point in given mesh to be exact)
probe points : 2.68655 -15.8401 20.9223 , 1.34332 -16.1237 19.8474
#-----------------
# Simulation time
#-----------------
# Times are given in milliseconds
PDE start time : 0.0
PDE end time : 25
#----------
# Geometry
#----------
# Meshes
2d mesh : ./meshes/atriaSurfacic
# The given mesh has units of cm, which is also CEPS length unit.
# So there is no need for scaling.
# geometry scale : 1
# Partitioning method can be
# - PTScotchNode
#--------
# Tissue
#--------
# Surface to volume ratio, in cm-1
Am : CONSTANT 200.0
# Surfacic membrane capacitance, in muF per cm2.
# If commented, Cm from the ionic model is used
# Cm : CONSTANT 1.0
# Conductivity, in mS per cm
intracellular conductivity : CONSTANT 1.2 0.4 0.4
extracellular conductivity : CONSTANT 1.2 0.4 0.4
#-------------
# Ionic model
#-------------
# Format: unknownIndex ionicModelName <regionAttributes>
# Here we use the Beeler-Reuter model everywhere
# for transmembrane voltage (component 0)
# No ionic model on extracellular potential
#-------------
# Stimulation
#-------------
# TIME options define the evolution of time of the stimulation.
# Here a one-time stim with C4 continuous profile that lasts 2 ms
# SPACE options define where the stimulation is applied.
# Here as well a C4 bell shaped stimulation, centered at 0.5 0.5 0 with
# 0.05 diameter
# Amplitude is in muA per cm2
stimulation: TIME PROFILE
C4 START 0.04 DURATION 2. \
SPACE PROFILE
C4 CENTER 0.5 0.5 0. DIAMETER 0.05 AMPLITUDE 5000
#----------------
# solver options
#----------------
# Only FE 1 available for now
spatial discretization : FE 1
# Time scheme for PDE solving
PDE solver : SBDF 2
PDE time step : 0.05
# Linear solver options
absolute tolerance : 1.E-12
relative tolerance : 1.E-12
solver type : GMRES
# Ionic solver. Here Rush Larsen of order 2.
ionic model solver : RL 2
@ PTScotchNode
GeomNode SCOTCH partitioning using primal graph.
Beeler Reuter (1977) ionic model.
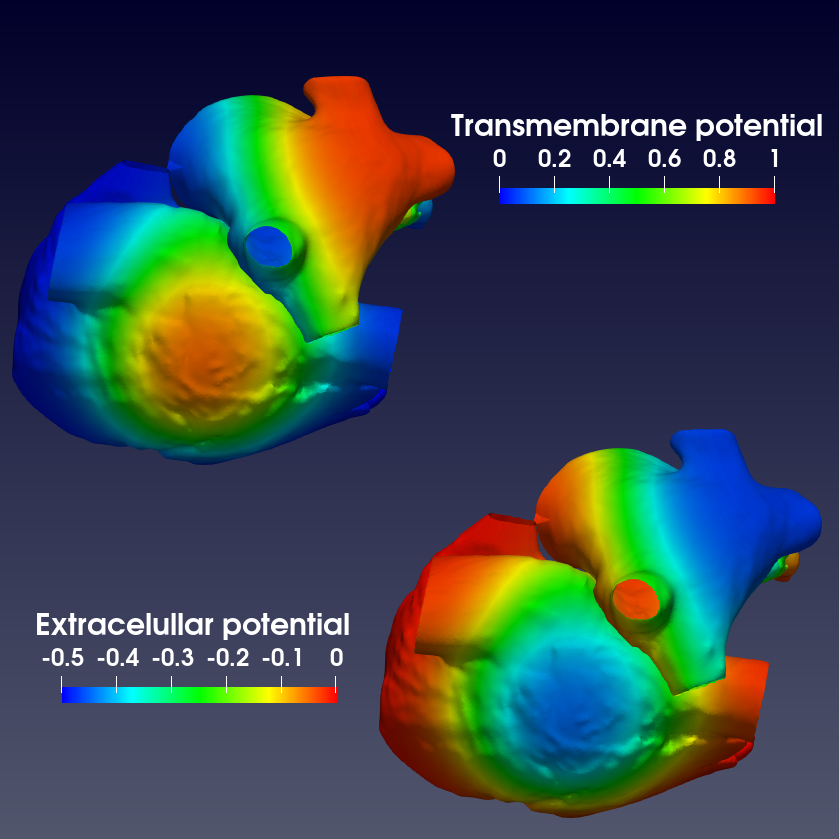
Here is an example of output you can get with this model.